F. Wayne Outten Group Site
Our lab seeks to define the mechanisms of iron (Fe) homeostasis in bacteria to disrupt
this essential process in bacterial pathogens to attenuate their growth in the human
host. We use the bacterium Escherichia coli as a model organism to study the fundamentals of in vivo iron metabolism.
Biological Chemistry of Bacterial Metal Metabolism
Fe-S clusters
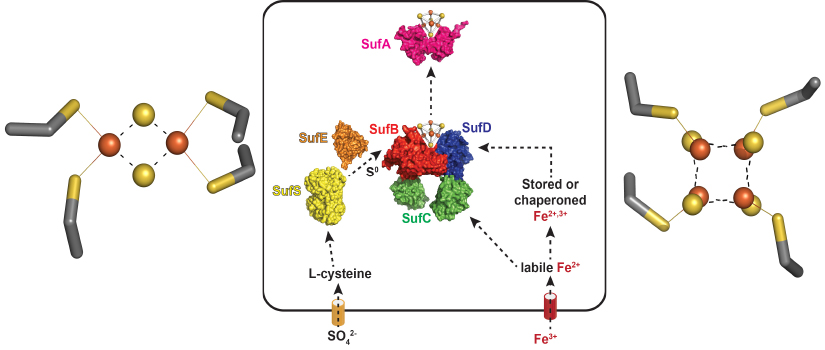
Fe-S clusters are metal cofactors required by proteins that carry out many of life's
most essential processes, such as respiration, nitrogen fixation, central carbon metabolism,
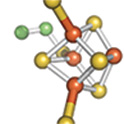
and DNA repair. For example, the left panel shows the [4Fe-4S] cluster of aconitase,
a key enzyme in the TCA cycle. Building and maintaining Fe-S clusters is a challenge
to most cells because of the scarcity of iron in the environment and due to the presence
of oxygen, which can react with and damage Fe-S clusters. Our NIH-funded project studies
a pathway in bacteria, know as Suf, that is used to build Fe-S clusters under harsh
conditions. A model of Suf function is shown in the banner above (center panel).

Bacteria
Single-celled bacteria can often serve as useful model organisms for molecular studies
because they are rapidly growing, easy to manipulate genetically, and often have genes
that are conserved in higher eukaryotes. We also study bacteria to understand their
physiological and biochemical differences from higher organisms (like humans) so that
we can exploit those differences to disrupt the survival of harmful bacterial pathogens.
Most of our previous studies were using the "workhorse" organism Escherichia coli
to study the fundamental details of iron homeostasis.
Contact Information
Office Location: GSRC 309
Office Phone: 803-777-8151
Lab Phone: 803-777-0669
outtenf@mailbox.sc.edu